MetaNetX Ontology
Release: 2017-11-13
- Modified on: 2024-05-14
- This version:
- http://purl.org/net/mnx/1.0
- Authors:
- Marco Pagni, https://www.sib.swiss✉
- Sébastien Moretti, https://www.sib.swiss✉
- Download serialization:
- License:
- Visualization:
- Cite as:
- MetaNetX/MNXref: unified namespace for metabolites and biochemical reactions in the context of metabolic models Sébastien Moretti, Van Du T Tran, Florence Mehl, Mark Ibberson, Marco Pagni Nucleic Acids Research (2021), 49(D1):D570-D574
Abstract
MetaNetX/MNXref is a reconciliation of metabolites and biochemical reactions, providing cross-links between major public biochemistry and Genome-Scale Metabolic Network (GSMN) databases.Introduction back to ToC
GSMN reconstruction is one of the foundations of Systems Biology. The development of such reconstruction involves the integration of knowledge about reactions and metabolites from the scientific literature, public databases and previously published GSMNs. Historically, many GSMNs were formulated using metabolites represented as "symbols", i.e. without explicit reference to a molecular structure. The initial motivation for creating this resource ten years ago was to add molecular structures to existing GSMNs. More generally, the goal was to establish cross-links between symbols in GSMNs published by different groups and the molecules found in the major biochemical databases. This problem is trivial as long as one-to-one mappings can be established between the different resources, but this is not always the case. Merging metabolites in a metabolic network may lead to the merging of reactions, possibly altering the model properties, and hence the predictions that could be made using it.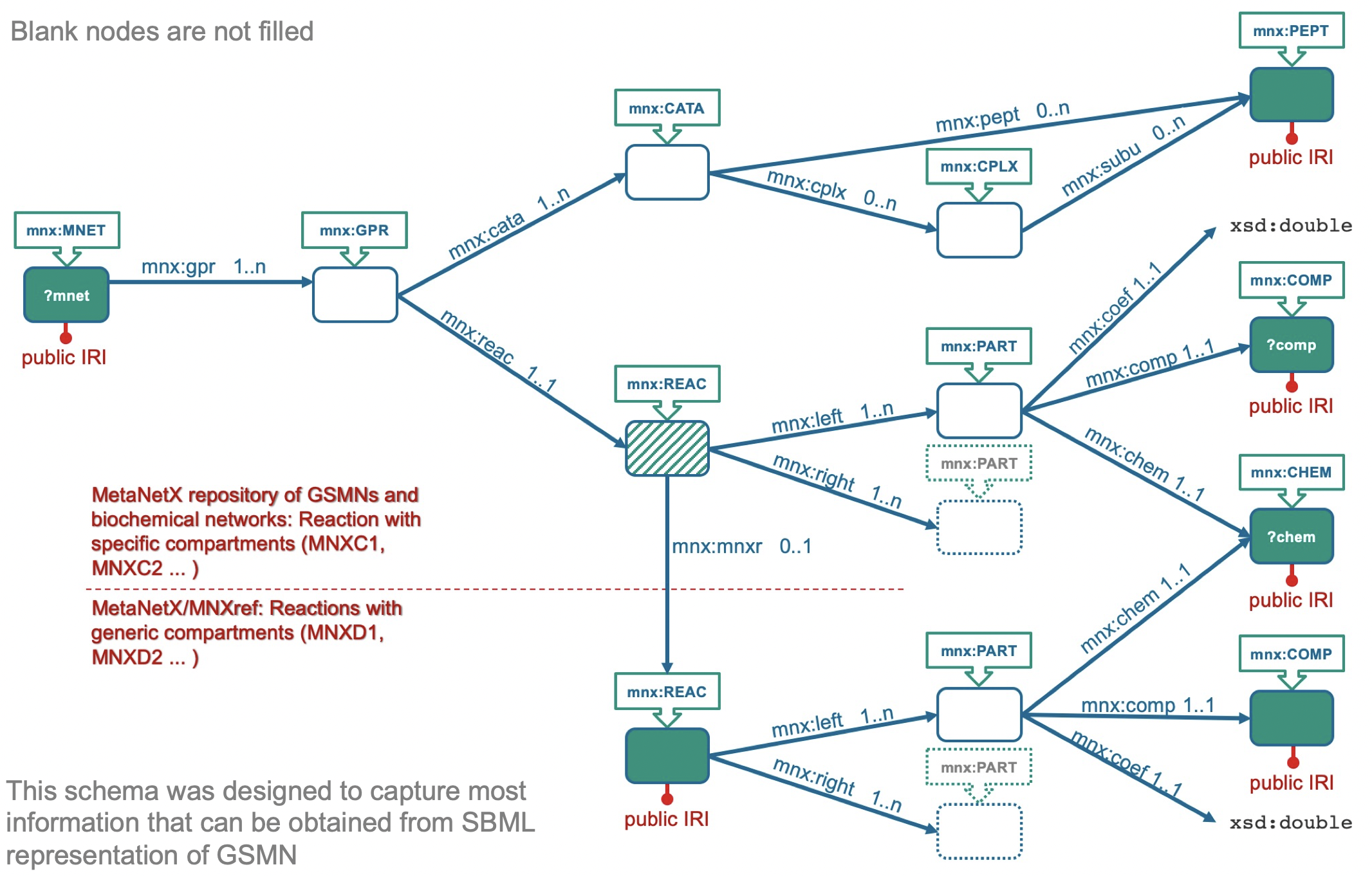
MetaNetX Ontology: Overview back to ToC
This ontology has the following classes and properties.Classes
- C A T A
- G P R
- MetaNetX compartment, i.e. a set of sub-cellular compartments
- MetaNetX metabolite class
- MetaNetX reaction, i.e. a set of chemical reaction
- mnx:EMPTY
- mnx:MNXR01
- P A R T
- P E P T
Object Properties
- cata
- chem Refer
- chem Replaced By
- chem Source
- comp Refer
- comp Replaced By
- comp Source
- comp Xref
- contains chem member
- cplx
- gene Name
- gpr
- has reference chem
- has stereo child
- left
- mnxr
- pept
- pept Xref
- reac
- reac Isom
- reac Replaced By
- reac Source
- reac Xref
- right
- side
- subu
Data Properties
- can Grow
- charge
- chem Count
- chem In M N Xref
- classification
- comp Count
- comp In M N Xref
- formula
- inchi
- inchikey
- is Balanced
- is Generic Comp
- is Transport
- lineage
- mass
- organism
- pept Count
- pmid
- reac Count
- reac In M N Xref
- smiles
- spec Count
- taxid
Named Individuals
MetaNetX Ontology: Description back to ToC
MetaNetX/MNXref: unified namespace for metabolites and biochemical reactions in the context of metabolic modelsCross-reference for MetaNetX Ontology classes, object properties and data properties back to ToC
This section provides details for each class and property defined by MetaNetX Ontology.Classes
- C A T A
- G P R
- MetaNetX compartment, i.e. a set of sub-cellular compartments
- MetaNetX metabolite class
- MetaNetX reaction, i.e. a set of chemical reaction
- mnx:EMPTY
- mnx:MNXR01
- P A R T
- P E P T
C A T Ac back to ToC or Class ToC
IRI: https://rdf.metanetx.org/schema/CATA
G P Rc back to ToC or Class ToC
IRI: https://rdf.metanetx.org/schema/GPR
MetaNetX compartment, i.e. a set of sub-cellular compartmentsc back to ToC or Class ToC
IRI: https://rdf.metanetx.org/schema/COMP
- is in domain of
- comp In M N Xref dp, comp Refer op, comp Replaced By op, comp Source op, comp Xref op, is Generic Comp dp
- is in range of
- comp Replaced By op
- has members
- mnx:BOUNDARY ni
MetaNetX metabolite classc back to ToC or Class ToC
IRI: https://rdf.metanetx.org/schema/CHEM
- is in domain of
- charge dp, chem In M N Xref dp, chem Refer op, chem Replaced By op, chem Source op, contains chem member op, formula dp, has stereo child op, inchi dp, inchikey dp, mass dp, smiles dp
- is in range of
- chem Replaced By op, has stereo child op
- has members
- mnx:BIOMASS ni, mnx:HYDROXYDE ni, mnx:PMF ni, mnx:PROTON ni, mnx:WATER ni
MetaNetX reaction, i.e. a set of chemical reactionc back to ToC or Class ToC
IRI: https://rdf.metanetx.org/schema/REAC
- is in domain of
- classification dp, has reference chem op, is Balanced dp, is Transport dp, left op, mnxr op, reac In M N Xref dp, reac Isom op, reac Replaced By op, reac Source op, reac Xref op, right op, side op
- is in range of
- mnxr op, reac op, reac Isom op, reac Replaced By op
mnx:EMPTY c back to ToC or Class ToC
IRI: https://rdf.metanetx.org/schema/EMPTY
mnx:MNXR01c back to ToC or Class ToC
IRI: https://rdf.metanetx.org/schema/MNXR01
Object Properties
- cata
- chem Refer
- chem Replaced By
- chem Source
- comp Refer
- comp Replaced By
- comp Source
- comp Xref
- contains chem member
- cplx
- gene Name
- gpr
- has reference chem
- has stereo child
- left
- mnxr
- pept
- pept Xref
- reac
- reac Isom
- reac Replaced By
- reac Source
- reac Xref
- right
- side
- subu
cataop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/cata
chem Referop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/chemRefer
- has domain
- MetaNetX metabolite class c
- has range
- Resource c
chem Replaced Byop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/chemReplacedBy
- has domain
- MetaNetX metabolite class c
- has range
- MetaNetX metabolite class c
chem Sourceop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/chemSource
- has domain
- MetaNetX metabolite class c
- has range
- Resource c
comp Referop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/compRefer
- has domain
- MetaNetX compartment, i.e. a set of sub-cellular compartments c
- has range
- Resource c
comp Replaced Byop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/compReplacedBy
comp Sourceop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/compSource
- has domain
- MetaNetX compartment, i.e. a set of sub-cellular compartments c
- has range
- Resource c
comp Xrefop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/compXref
- has domain
- MetaNetX compartment, i.e. a set of sub-cellular compartments c
- has range
- Resource c
contains chem memberop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/chemXref
- has domain
- MetaNetX metabolite class c
- has range
- Resource c
cplxop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/cplx
gene Nameop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/geneName
gprop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/gpr
has reference chemop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/reacRefer
- has domain
- MetaNetX reaction, i.e. a set of chemical reaction c
- has range
- Resource c
has stereo childop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/chemIsom
- has domain
- MetaNetX metabolite class c
- has range
- MetaNetX metabolite class c
- is same as
- has Isomeric Child ni
- is also defined as
- named individual
leftop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/left
- has domain
- MetaNetX reaction, i.e. a set of chemical reaction c
- has range
- P A R T c
mnxrop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/mnxr
- has domain
- MetaNetX reaction, i.e. a set of chemical reaction c
- has range
- MetaNetX reaction, i.e. a set of chemical reaction c
peptop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/pept
pept Xrefop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/peptXref
reacop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/reac
- has domain
- G P R c
- has range
- MetaNetX reaction, i.e. a set of chemical reaction c
reac Isomop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/reacIsom
- has domain
- MetaNetX reaction, i.e. a set of chemical reaction c
- has range
- MetaNetX reaction, i.e. a set of chemical reaction c
- is also defined as
- annotation property
reac Replaced Byop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/reacReplacedBy
- has domain
- MetaNetX reaction, i.e. a set of chemical reaction c
- has range
- MetaNetX reaction, i.e. a set of chemical reaction c
reac Sourceop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/reacSource
- has domain
- MetaNetX reaction, i.e. a set of chemical reaction c
- has range
- Resource c
reac Xrefop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/reacXref
- has domain
- MetaNetX reaction, i.e. a set of chemical reaction c
- has range
- Resource c
rightop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/right
- has domain
- MetaNetX reaction, i.e. a set of chemical reaction c
- has range
- P A R T c
sideop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/side
- has domain
- MetaNetX reaction, i.e. a set of chemical reaction c
- has range
- P A R T c
subuop back to ToC or Object Property ToC
IRI: https://rdf.metanetx.org/schema/subu
Data Properties
- can Grow
- charge
- chem Count
- chem In M N Xref
- classification
- comp Count
- comp In M N Xref
- formula
- inchi
- inchikey
- is Balanced
- is Generic Comp
- is Transport
- lineage
- mass
- organism
- pept Count
- pmid
- reac Count
- reac In M N Xref
- smiles
- spec Count
- taxid
can Growdp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/canGrow
chargedp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/charge
- has domain
- MetaNetX metabolite class c
- has range
- integer
chem Countdp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/chemCount
chem In M N Xrefdp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/chemInMNXref
- has domain
- MetaNetX metabolite class c
- has range
- boolean
classificationdp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/classification
- has domain
- MetaNetX reaction, i.e. a set of chemical reaction c
- has range
- string
comp Countdp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/compCount
comp In M N Xrefdp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/compInMNXref
- has domain
- MetaNetX compartment, i.e. a set of sub-cellular compartments c
- has range
- boolean
formuladp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/formula
- has domain
- MetaNetX metabolite class c
- has range
- string
inchidp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/inchi
- has domain
- MetaNetX metabolite class c
- has range
- string
inchikeydp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/inchikey
- has domain
- MetaNetX metabolite class c
- has range
- string
is Balanceddp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/isBalanced
- has domain
- MetaNetX reaction, i.e. a set of chemical reaction c
- has range
- boolean
is Generic Compdp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/isGenericComp
- has domain
- MetaNetX compartment, i.e. a set of sub-cellular compartments c
- has range
- boolean
is Transportdp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/isTransport
- has domain
- MetaNetX reaction, i.e. a set of chemical reaction c
- has range
- boolean
lineagedp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/lineage
massdp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/mass
- has domain
- MetaNetX metabolite class c
- has range
- decimal
organismdp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/organism
pept Countdp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/peptCount
pmiddp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/pmid
- has range
- M N E T c
reac Countdp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/reacCount
reac In M N Xrefdp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/reacInMNXref
- has domain
- MetaNetX reaction, i.e. a set of chemical reaction c
- has range
- boolean
smilesdp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/smiles
- has domain
- MetaNetX metabolite class c
- has range
- string
spec Countdp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/specCount
taxiddp back to ToC or Data Property ToC
IRI: https://rdf.metanetx.org/schema/taxid
- has range
- M N E T c
Named Individuals
- has stereo child
- Marco Pagni
- mnx:BIOMASS
- mnx:BOUNDARY
- mnx:HYDROXYDE
- mnx:PMF
- mnx:PROTON
- mnx:SPONTANEOUS
- mnx:WATER
- Sébastien Moretti
has stereo childni back to ToC or Named Individual ToC
IRI: https://rdf.metanetx.org/schema/chemIsom
- is same as
- has Isomeric Child ni
- has facts
- same As ep has Isomeric Child ni
- is also defined as
- object property
Marco Pagnini back to ToC or Named Individual ToC
IRI: https://orcid.org/0000-0001-9292-9463
- belongs to
- Person c
- Person c
- has facts
- affiliation ap https://www.sib.swiss ep
- email ap mailto:marco.pagni@sib.swiss ep
- name ap "Marco Pagni"
mnx:BIOMASSni back to ToC or Named Individual ToC
IRI: https://rdf.metanetx.org/schema/BIOMASS
- belongs to
- MetaNetX metabolite class c
mnx:BOUNDARYni back to ToC or Named Individual ToC
IRI: https://rdf.metanetx.org/schema/BOUNDARY
mnx:HYDROXYDEni back to ToC or Named Individual ToC
IRI: https://rdf.metanetx.org/schema/HYDROXYDE
- belongs to
- MetaNetX metabolite class c
mnx:PMFni back to ToC or Named Individual ToC
IRI: https://rdf.metanetx.org/schema/PMF
- belongs to
- MetaNetX metabolite class c
mnx:PROTONni back to ToC or Named Individual ToC
IRI: https://rdf.metanetx.org/schema/PROTON
- belongs to
- MetaNetX metabolite class c
mnx:SPONTANEOUSni back to ToC or Named Individual ToC
IRI: https://rdf.metanetx.org/schema/SPONTANEOUS
- belongs to
- P E P T c
mnx:WATERni back to ToC or Named Individual ToC
IRI: https://rdf.metanetx.org/schema/WATER
- belongs to
- MetaNetX metabolite class c
Sébastien Morettini back to ToC or Named Individual ToC
IRI: https://orcid.org/0000-0003-3947-488X
- belongs to
- Person c
- Person c
- has facts
- affiliation ap https://www.sib.swiss ep
- email ap mailto:Sebastien.Moretti@sib.swiss ep
- name ap "Sébastien Moretti"
Legend back to ToC
op: Object Properties
dp: Data Properties
ni: Named Individuals
ep: External Properties
References back to ToC
Add your references here. It is recommended to have them as a list.Acknowledgments back to ToC
The authors would like to thank Silvio Peroni for developing LODE, a Live OWL Documentation Environment, which is used for representing the Cross Referencing Section of this document and Daniel Garijo for developing Widoco, the program used to create the template used in this documentation.





